Tuesday, December 13, 2016
Thursday, November 24, 2016
To create a new app go to https://developers.facebook.com
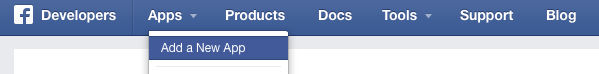
Click on “Apps” and choose “Add a New App“. In the next window choose “Website” and give your app a fancy name.
After clicking on “Create a New App ID“, choose a category for your app in the next window and apply the changes with “Create App ID“.
You can then click on “Skip Quick Start” to get directly to the settings of your app.

Welcome to your first own Facebook app!
R
First we need to install the packages devtools and Rfacebook from github as this is currently the most recent version.
1
2
|
install.packages("devtools")
library(devtools)
|
1
|
install_github("Rfacebook", "pablobarbera", subdir="Rfacebook")
|
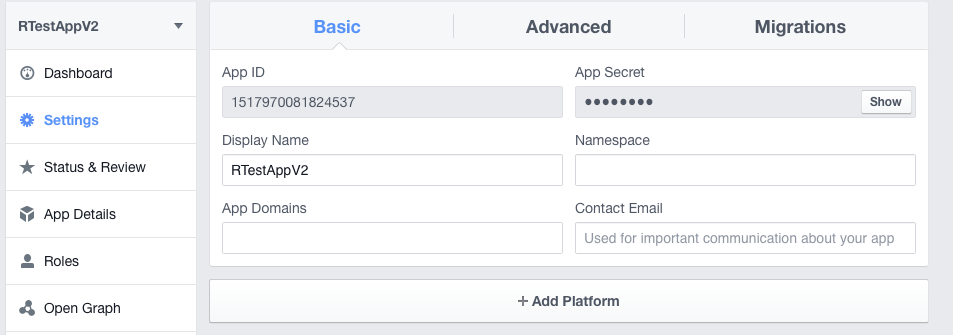
Just copy your app id and your app secret from your app settings on the Facebook developer page.
1
2
3
|
require("Rfacebook")
fb_oauth <- fbOAuth(app_id="123456789", app_secret="1A2B3C4D",extended_permissions = TRUE)
|
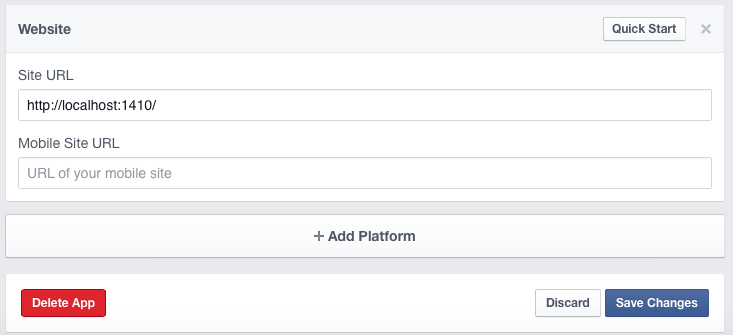
Copy the URL and go to the settings of your Facebook app. Click on the settings tab on the left side and then choose “+ Add Platform“.
Then add the URL in the field “Site URL” and save the changes.
Go back to your R session and hit enter. Then a browser window should open you have to allow the app to access your Facebook account.
If everything worked the browser should show the message
And your R console will confirm it with
You can then save your fb_oauth object and use it for the next time.
1
2
3
|
save(fb_oauth, file="fb_oauth")
load("fb_oauth")
|
Analyze Facebook with R!
Now we connected everything and have access to Facebook. We will start with getting our own profile information.As I mentioned before because of Facebook´s new API policies the information you can get is very limited compared to the amount you were able to download with apps using API 1.
So I will just show you now how to get your own personal information but other posts will follow with new use-cases of the new API version.
The getUsers function returns public information about one or more Facebook user. If we use “me” as the username argument, it will return our own profile info.
1
|
me <- getUsers("me",token=fb_oauth)
|
An advantage of the new API version is that you can get more than 100 likes. You can get the things you liked with:
1
|
my_likes <- getLikes(user="me", token=fb_oauth)
|
=========================================face-book ======================================================================================
install.packages("devtools")
library(devtools)
library(httr)
set_config(
use_proxy(url="proxy.company.co.in", port=8080, username="user@company.co.in",password="password@4321")
)
install_github("Rfacebook", "pablobarbera", subdir="Rfacebook")
require("Rfacebook")
fb_oauth <- fbOAuth(app_id="331", app_secret="f82e0ffc352f4b30c16686ccf",extended_permissions = TRUE)
save(fb_oauth, file="fb_oauth")
load("fb_oauth")
axis_page <- getPage(page="axisbank", token=fb_oauth)
icici_page <- getPage(page="icicibank", token=fb_oauth)
fb_page <- getPage(page="facebook", token=fb_oauth)
token <- "EAACEdEose0cBA"
my_friends <- getFriends(token=token, simplify=TRUE)
Wednesday, November 23, 2016
Spark submit with SBT
Spark submit with SBT
This recipe assumes sbt is installed and you have already gone over mysql with Spark recipe.I am a big fan of Spark Shell. Biggest proof is Spark Cookbook which has all recipes in the form of collection of single commands on Spark Shell. It makes it easy to understand and run and see what exact effect each command is having.
On similar lines, I have a big fan of Maven. Maven brought two disruptive changes to the build world which are going to stay forever and they are
- Declarative dependency management
- Convention over configuration
spark-shell is not enough and we have to use spark-submit.
spark-submit expects the application logic to be bundled in a jar file.
Now creating this jar file using maven is a lot of work especially for
super simple project and this is where simplicity of Sbt comes into the
picture. In this recipe we’ll build and deploy a simple application using Sbt.
Create directory people and src/main/scala in that.
$ mkdir -p people/src/main/scala |
import org.apache.spark._ import org.apache.spark.sql._ object Person extends App { val sc = new SparkContext val sqlContext = new SQLContext(sc) val url="jdbc:mysql://localhost:3306/hadoopdb" val prop = new java.util.Properties prop.setProperty("user","hduser") prop.setProperty("password","vipassana") val people = sqlContext.read.jdbc(url,"person",prop) people.collect.foreach(println) } |
$ echo "libraryDependencies += \"org.apache.spark\" %% \"spark-sql\" % \"1.4.1\"" >> build.sbt |
$ sbt clean package
|
$ spark-submit --driver-class-path /home/hduser/thirdparty/mysql.jar target/scala-2.10/people*.jar |
Now let’s do the same using cluster deploy mode, in this case we have to go to http://spark-master:8080 to see the results.
$ spark-submit --master spark://localhost:7077 --deploy-mode cluster --driver-class-path /home/hduser/thirdparty/mysql.jar --jars /home/hduser/thirdparty/mysql.jar target/scala-2.10/people*.jar |
Tuesday, November 8, 2016
HBase shell commands
HBase shell commands
HBase shell commands are mainly categorized into 6 parts
1) General HBase shell commands
| status | Show cluster status. Can be ‘summary’, ‘simple’, or ‘detailed’. The default is ‘summary’. hbase> status hbase> status ‘simple’ hbase> status ‘summary’ hbase> status ‘detailed’ |
| version | Output this HBase versionUsage: hbase> version |
| whoami | Show the current hbase user.Usage: hbase> whoami |
| alter | Alter column family schema; pass table name and a dictionary specifying new column family schema. Dictionaries are described on the main help command output. Dictionary must include name of column family to alter.For example, to change or add the ‘f1’ column family in table ‘t1’ from current value to keep a maximum of 5 cell VERSIONS, do: hbase> alter ‘t1’, NAME => ‘f1’, VERSIONS => 5 You can operate on several column families: hbase> alter ‘t1’, ‘f1’, {NAME => ‘f2’, IN_MEMORY => true}, {NAME => ‘f3’, VERSIONS => 5} To delete the ‘f1’ column family in table ‘t1’, use one of:hbase> alter ‘t1’, NAME => ‘f1’, METHOD => ‘delete’ hbase> alter ‘t1’, ‘delete’ => ‘f1’ You can also change table-scope attributes like MAX_FILESIZE, READONLY, MEMSTORE_FLUSHSIZE, DEFERRED_LOG_FLUSH, etc. These can be put at the end; for example, to change the max size of a region to 128MB, do: hbase> alter ‘t1’, MAX_FILESIZE => ‘134217728’ You can add a table coprocessor by setting a table coprocessor attribute: hbase> alter ‘t1’, ‘coprocessor’=>’hdfs:///foo.jar|com.foo.FooRegionObserver|1001|arg1=1,arg2=2’ Since you can have multiple coprocessors configured for a table, a sequence number will be automatically appended to the attribute name to uniquely identify it. The coprocessor attribute must match the pattern below in order for the framework to understand how to load the coprocessor classes: [coprocessor jar file location] | class name | [priority] | [arguments] You can also set configuration settings specific to this table or column family: hbase> alter ‘t1’, CONFIGURATION => {‘hbase.hregion.scan.loadColumnFamiliesOnDemand’ => ‘true’} hbase> alter ‘t1’, {NAME => ‘f2’, CONFIGURATION => {‘hbase.hstore.blockingStoreFiles’ => ’10’}} You can also remove a table-scope attribute: hbase> alter ‘t1’, METHOD => ‘table_att_unset’, NAME => ‘MAX_FILESIZE’ hbase> alter ‘t1’, METHOD => ‘table_att_unset’, NAME => ‘coprocessor$1’ There could be more than one alteration in one command: hbase> alter ‘t1’, { NAME => ‘f1’, VERSIONS => 3 }, { MAX_FILESIZE => ‘134217728’ }, { METHOD => ‘delete’, NAME => ‘f2’ }, OWNER => ‘johndoe’, METADATA => { ‘mykey’ => ‘myvalue’ } |
| create | Create table; pass table name, a dictionary of specifications per column family, and optionally a dictionary of table configuration. hbase> create ‘t1’, {NAME => ‘f1’, VERSIONS => 5} hbase> create ‘t1’, {NAME => ‘f1’}, {NAME => ‘f2’}, {NAME => ‘f3’} hbase> # The above in shorthand would be the following: hbase> create ‘t1’, ‘f1’, ‘f2’, ‘f3’ hbase> create ‘t1’, {NAME => ‘f1’, VERSIONS => 1, TTL => 2592000, BLOCKCACHE => true} hbase> create ‘t1’, {NAME => ‘f1’, CONFIGURATION => {‘hbase.hstore.blockingStoreFiles’ => ’10’}} Table configuration options can be put at the end. |
| describe | Describe the named table. hbase> describe ‘t1’ |
| disable | Start disable of named table hbase> disable ‘t1’ |
| disable_all | Disable all of tables matching the given regex hbase> disable_all ‘t.*’ |
| is_disabled | verifies Is named table disabled hbase> is_disabled ‘t1’ |
| drop | Drop the named table. Table must first be disabled hbase> drop ‘t1’ |
| drop_all | Drop all of the tables matching the given regex hbase> drop_all ‘t.*’ |
| enable | Start enable of named table hbase> enable ‘t1’ |
| enable_all | Enable all of the tables matching the given regex hbase> enable_all ‘t.*’ |
| is_enabled | verifies Is named table enabled hbase> is_enabled ‘t1’ |
| exists | Does the named table exist hbase> exists ‘t1’ |
| list | List all tables in hbase. Optional regular expression parameter could be used to filter the output hbase> list hbase> list ‘abc.*’ |
| show_filters | Show all the filters in hbase. hbase> show_filters |
| alter_status | Get the status of the alter command. Indicates the number of regions
of the table that have received the updated schema Pass table name. hbase> alter_status ‘t1’ |
| alter_async | Alter column family schema, does not wait for all regions to receive the schema changes. Pass table name and a dictionary specifying new column family schema. Dictionaries are described on the main help command output. Dictionary must include name of column family to alter. To change or add the ‘f1’ column family in table ‘t1’ from defaults to instead keep a maximum of 5 cell VERSIONS, do:hbase> alter_async ‘t1’, NAME => ‘f1’, VERSIONS => 5To delete the ‘f1’ column family in table ‘t1’, do: hbase> alter_async ‘t1’, NAME => ‘f1’, METHOD => ‘delete’or a shorter version:hbase> alter_async ‘t1’, ‘delete’ => ‘f1’ You can also change table-scope attributes like MAX_FILESIZE MEMSTORE_FLUSHSIZE, READONLY, and DEFERRED_LOG_FLUSH. For example, to change the max size of a family to 128MB, do: hbase> alter ‘t1’, METHOD => ‘table_att’, MAX_FILESIZE => ‘134217728’ There could be more than one alteration in one command: hbase> alter ‘t1’, {NAME => ‘f1’}, {NAME => ‘f2’, METHOD => ‘delete’} To check if all the regions have been updated, use alter_status <table_name> |
| count | Count the number of rows in a table. Return value is the number of rows. This operation may take a LONG time (Run ‘$HADOOP_HOME/bin/hadoop jar hbase.jar rowcount’ to run a counting mapreduce job). Current count is shown every 1000 rows by default. Count interval may be optionally specified. Scan caching is enabled on count scans by default. Default cache size is 10 rows. If your rows are small in size, you may want to increase this parameter. Examples:hbase> count ‘t1’ hbase> count ‘t1’, INTERVAL => 100000 hbase> count ‘t1’, CACHE => 1000 hbase> count ‘t1’, INTERVAL => 10, CACHE => 1000 The same commands also can be run on a table reference. Suppose you had a reference t to table ‘t1’, the corresponding commands would be:hbase> t.count hbase> t.count INTERVAL => 100000 hbase> t.count CACHE => 1000 hbase> t.count INTERVAL => 10, CACHE => 1000 |
| delete | Put a delete cell value at specified table/row/column and optionally timestamp coordinates. Deletes must match the deleted cell’s coordinates exactly. When scanning, a delete cell suppresses older versions. To delete a cell from ‘t1’ at row ‘r1’ under column ‘c1’ marked with the time ‘ts1’, do: hbase> delete ‘t1’, ‘r1’, ‘c1’, ts1 The same command can also be run on a table reference. Suppose you had a reference t to table ‘t1’, the corresponding command would be:hbase> t.delete ‘r1’, ‘c1’, ts1 |
| deleteall | Delete all cells in a given row; pass a table name, row, and optionally a column and timestamp. Examples:hbase> deleteall ‘t1’, ‘r1’ hbase> deleteall ‘t1’, ‘r1’, ‘c1’ hbase> deleteall ‘t1’, ‘r1’, ‘c1’, ts1 The same commands also can be run on a table reference. Suppose you had a reference t to table ‘t1’, the corresponding command would be:hbase> t.deleteall ‘r1’ hbase> t.deleteall ‘r1’, ‘c1’ hbase> t.deleteall ‘r1’, ‘c1’, ts1 |
| get | Get row or cell contents; pass table name, row, and optionally a dictionary of column(s), timestamp, timerange and versions. Examples: hbase> get ‘t1’, ‘r1’ hbase> get ‘t1’, ‘r1’, {TIMERANGE => [ts1, ts2]} hbase> get ‘t1’, ‘r1’, {COLUMN => ‘c1’} hbase> get ‘t1’, ‘r1’, {COLUMN => [‘c1’, ‘c2’, ‘c3’]} hbase> get ‘t1’, ‘r1’, {COLUMN => ‘c1’, TIMESTAMP => ts1} hbase> get ‘t1’, ‘r1’, {COLUMN => ‘c1’, TIMERANGE => [ts1, ts2], VERSIONS => 4} hbase> get ‘t1’, ‘r1’, {COLUMN => ‘c1’, TIMESTAMP => ts1, VERSIONS => 4} hbase> get ‘t1’, ‘r1’, {FILTER => “ValueFilter(=, ‘binary:abc’)”} hbase> get ‘t1’, ‘r1’, ‘c1’ hbase> get ‘t1’, ‘r1’, ‘c1’, ‘c2’ hbase> get ‘t1’, ‘r1’, [‘c1’, ‘c2’] Besides the default ‘toStringBinary’ format, ‘get’ also supports custom formatting by column. A user can define a FORMATTER by adding it to the column name in the get specification. The FORMATTER can be stipulated:1. either as a org.apache.hadoop.hbase.util.Bytes method name (e.g, toInt, toString) 2. or as a custom class followed by method name: e.g. ‘c(MyFormatterClass).format’.Example formatting cf:qualifier1 and cf:qualifier2 both as Integers: hbase> get ‘t1’, ‘r1’ {COLUMN => [‘cf:qualifier1:toInt’, ‘cf:qualifier2:c(org.apache.hadoop.hbase.util.Bytes).toInt’] } Note that you can specify a FORMATTER by column only (cf:qualifer). You cannot specify a FORMATTER for all columns of a column family.The same commands also can be run on a reference to a table (obtained via get_table or create_table). Suppose you had a reference t to table ‘t1’, the corresponding commands would be: hbase> t.get ‘r1’ hbase> t.get ‘r1’, {TIMERANGE => [ts1, ts2]} hbase> t.get ‘r1’, {COLUMN => ‘c1’} hbase> t.get ‘r1’, {COLUMN => [‘c1’, ‘c2’, ‘c3’]} hbase> t.get ‘r1’, {COLUMN => ‘c1’, TIMESTAMP => ts1} hbase> t.get ‘r1’, {COLUMN => ‘c1’, TIMERANGE => [ts1, ts2], VERSIONS => 4} hbase> t.get ‘r1’, {COLUMN => ‘c1’, TIMESTAMP => ts1, VERSIONS => 4} hbase> t.get ‘r1’, {FILTER => “ValueFilter(=, ‘binary:abc’)”} hbase> t.get ‘r1’, ‘c1’ hbase> t.get ‘r1’, ‘c1’, ‘c2’ hbase> t.get ‘r1’, [‘c1’, ‘c2’] |
| get_counter | Return a counter cell value at specified table/row/column coordinates. A cell cell should be managed with atomic increment function oh HBase and the data should be binary encoded. Example: hbase> get_counter ‘t1’, ‘r1’, ‘c1’ The same commands also can be run on a table reference. Suppose you had a reference t to table ‘t1’, the corresponding command would be: hbase> t.get_counter ‘r1’, ‘c1’ |
| incr | Increments a cell ‘value’ at specified table/row/column coordinates. To increment a cell value in table ‘t1’ at row ‘r1’ under column ‘c1’ by 1 (can be omitted) or 10 do: hbase> incr ‘t1’, ‘r1’, ‘c1’ hbase> incr ‘t1’, ‘r1’, ‘c1’, 1 hbase> incr ‘t1’, ‘r1’, ‘c1’, 10 The same commands also can be run on a table reference. Suppose you had a reference t to table ‘t1’, the corresponding command would be:hbase> t.incr ‘r1’, ‘c1’ hbase> t.incr ‘r1’, ‘c1’, 1 hbase> t.incr ‘r1’, ‘c1’, 10 |
| put | Put a cell ‘value’ at specified table/row/column and optionally timestamp coordinates. To put a cell value into table ‘t1’ at row ‘r1’ under column ‘c1’ marked with the time ‘ts1’, do: hbase> put ‘t1’, ‘r1’, ‘c1’, ‘value’, ts1 The same commands also can be run on a table reference. Suppose you had a reference t to table ‘t1’, the corresponding command would be: hbase> t.put ‘r1’, ‘c1’, ‘value’, ts1 |
| scan | Scan a table; pass table name and optionally a dictionary of scanner specifications. Scanner specifications may include one or more of: TIMERANGE, FILTER, LIMIT, STARTROW, STOPROW, TIMESTAMP, MAXLENGTH, or COLUMNS, CACHEIf no columns are specified, all columns will be scanned. To scan all members of a column family, leave the qualifier empty as in ‘col_family:’.The filter can be specified in two ways: 1. Using a filterString – more information on this is available in the Filter Language document attached to the HBASE-4176 JIRA 2. Using the entire package name of the filter.Some examples:hbase> scan ‘.META.’ hbase> scan ‘.META.’, {COLUMNS => ‘info:regioninfo’} hbase> scan ‘t1’, {COLUMNS => [‘c1’, ‘c2’], LIMIT => 10, STARTROW => ‘xyz’} hbase> scan ‘t1’, {COLUMNS => ‘c1’, TIMERANGE => [1303668804, 1303668904]} hbase> scan ‘t1’, {FILTER => “(PrefixFilter (‘row2’) AND (QualifierFilter (>=, ‘binary:xyz’))) AND (TimestampsFilter ( 123, 456))”} hbase> scan ‘t1’, {FILTER => org.apache.hadoop.hbase.filter.ColumnPaginationFilter.new(1, 0)} For experts, there is an additional option — CACHE_BLOCKS — which switches block caching for the scanner on (true) or off (false). By default it is enabled. Examples:hbase> scan ‘t1’, {COLUMNS => [‘c1’, ‘c2’], CACHE_BLOCKS => false} Also for experts, there is an advanced option — RAW — which instructs the scanner to return all cells (including delete markers and uncollected deleted cells). This option cannot be combined with requesting specific COLUMNS. Disabled by default. Example: hbase> scan ‘t1’, {RAW => true, VERSIONS => 10} Besides the default ‘toStringBinary’ format, ‘scan’ supports custom formatting by column. A user can define a FORMATTER by adding it to the column name in the scan specification. The FORMATTER can be stipulated: 1. either as a org.apache.hadoop.hbase.util.Bytes method name (e.g, toInt, toString) 2. or as a custom class followed by method name: e.g. ‘c(MyFormatterClass).format’. Example formatting cf:qualifier1 and cf:qualifier2 both as Integers: hbase> scan ‘t1’, {COLUMNS => [‘cf:qualifier1:toInt’, ‘cf:qualifier2:c(org.apache.hadoop.hbase.util.Bytes).toInt’] } Note that you can specify a FORMATTER by column only (cf:qualifer). You cannot specify a FORMATTER for all columns of a column family. Scan can also be used directly from a table, by first getting a reference to a table, like such: hbase> t = get_table ‘t’ hbase> t.scan Note in the above situation, you can still provide all the filtering, columns, options, etc as described above. |
| truncate | Disables, drops and recreates the specified table. Examples: hbase>truncate ‘t1’ |
| assign | Assign a region. Use with caution. If region already assigned, this command will do a force reassign. For experts only. Examples: hbase> assign ‘REGION_NAME’ |
| balancer | Trigger the cluster balancer. Returns true if balancer ran and was able to tell the region servers to unassign all the regions to balance (the re-assignment itself is async). Otherwise false (Will not run if regions in transition). Examples: hbase> balancer |
| balance_switch | Enable/Disable balancer. Returns previous balancer state. Examples: hbase> balance_switch true hbase> balance_switch false |
| close_region | Close a single region. Ask the master to close a region out on the cluster or if ‘SERVER_NAME’ is supplied, ask the designated hosting regionserver to close the region directly. Closing a region, the master expects ‘REGIONNAME’ to be a fully qualified region name. When asking the hosting regionserver to directly close a region, you pass the regions’ encoded name only. A region name looks like this:TestTable,0094429456,1289497600452.527db22f95c8a9e0116f0cc13c680396.The trailing period is part of the regionserver name. A region’s encoded name is the hash at the end of a region name; e.g. 527db22f95c8a9e0116f0cc13c680396 (without the period). A ‘SERVER_NAME’ is its host, port plus startcode. For example: host187.example.com,60020,1289493121758 (find servername in master ui or when you do detailed status in shell). This command will end up running close on the region hosting regionserver. The close is done without the master’s involvement (It will not know of the close). Once closed, region will stay closed. Use assign to reopen/reassign. Use unassign or move to assign the region elsewhere on cluster. Use with caution. For experts only. Examples:hbase> close_region ‘REGIONNAME’ hbase> close_region ‘REGIONNAME’, ‘SERVER_NAME’ |
| compact | Compact all regions in passed table or pass a region row to compact an individual region. You can also compact a single column family within a region. Examples: Compact all regions in a table: hbase> compact ‘t1’ Compact an entire region: hbase> compact ‘r1’ Compact only a column family within a region: hbase> compact ‘r1’, ‘c1’ Compact a column family within a table: hbase> compact ‘t1’, ‘c1’ |
| flush | Flush all regions in passed table or pass a region row to flush an individual region. For example:hbase> flush ‘TABLENAME’ hbase> flush ‘REGIONNAME’ |
| major_compact | Run major compaction on passed table or pass a region row to major compact an individual region. To compact a single column family within a region specify the region name followed by the column family name. Examples: Compact all regions in a table: hbase> major_compact ‘t1’ Compact an entire region: hbase> major_compact ‘r1’ Compact a single column family within a region: hbase> major_compact ‘r1’, ‘c1’ Compact a single column family within a table: hbase> major_compact ‘t1’, ‘c1’ |
| move | Move a region. Optionally specify target regionserver else we choose one at random. NOTE: You pass the encoded region name, not the region name so this command is a little different to the others. The encoded region name is the hash suffix on region names: e.g. if the region name were TestTable,0094429456,1289497600452.527db22f95c8a9e0116f0cc13c680396. then the encoded region name portion is 527db22f95c8a9e0116f0cc13c680396 A server name is its host, port plus startcode. For example: host187.example.com,60020,1289493121758 Examples:hbase> move ‘ENCODED_REGIONNAME’ hbase> move ‘ENCODED_REGIONNAME’, ‘SERVER_NAME’ |
| split | Split entire table or pass a region to split individual region. With the second parameter, you can specify an explicit split key for the region. Examples: split ‘tableName’ split ‘regionName’ # format: ‘tableName,startKey,id’ split ‘tableName’, ‘splitKey’ split ‘regionName’, ‘splitKey’ |
| unassign | Unassign a region. Unassign will close region in current location and then reopen it again. Pass ‘true’ to force the unassignment (‘force’ will clear all in-memory state in master before the reassign. If results in double assignment use hbck -fix to resolve. To be used by experts). Use with caution. For expert use only. Examples:hbase> unassign ‘REGIONNAME’ hbase> unassign ‘REGIONNAME’, true |
| hlog_roll | Roll the log writer. That is, start writing log messages to a new file. The name of the regionserver should be given as the parameter. A ‘server_name’ is the host, port plus startcode of a regionserver. For example: host187.example.com,60020,1289493121758 (find servername in master ui or when you do detailed status in shell) hbase>hlog_roll |
| zk_dump | Dump status of HBase cluster as seen by ZooKeeper. Example: hbase>zk_dump |
| add_peer | Add a peer cluster to replicate to, the id must be a short and the cluster key is composed like this: hbase.zookeeper.quorum:hbase.zookeeper.property.clientPort:zookeeper.znode.parent This gives a full path for HBase to connect to another cluster. Examples:hbase> add_peer ‘1’, “server1.cie.com:2181:/hbase” hbase> add_peer ‘2’, “zk1,zk2,zk3:2182:/hbase-prod” |
| remove_peer | Stops the specified replication stream and deletes all the meta information kept about it. Examples: hbase> remove_peer ‘1’ |
| list_peers | List all replication peer clusters. hbase> list_peers |
| enable_peer | Restarts the replication to the specified peer cluster, continuing from where it was disabled.Examples: hbase> enable_peer ‘1’ |
| disable_peer | Stops the replication stream to the specified cluster, but still keeps track of new edits to replicate.Examples: hbase> disable_peer ‘1’ |
| start_replication | Restarts all the replication features. The state in which each stream starts in is undetermined. WARNING: start/stop replication is only meant to be used in critical load situations. Examples: hbase> start_replication |
| stop_replication | Stops all the replication features. The state in which each stream stops in is undetermined. WARNING: start/stop replication is only meant to be used in critical load situations. Examples: hbase> stop_replication |
| grant | Grant users specific rights. Syntax : grantpermissions is either zero or more letters from the set “RWXCA”. READ(‘R’), WRITE(‘W’), EXEC(‘X’), CREATE(‘C’), ADMIN(‘A’)For example:hbase> grant ‘bobsmith’, ‘RWXCA’ hbase> grant ‘bobsmith’, ‘RW’, ‘t1’, ‘f1’, ‘col1’ |
| revoke | Revoke a user’s access rights. Syntax : revoke For example: hbase> revoke ‘bobsmith’, ‘t1’, ‘f1’, ‘col1’ |
| user_permission | Show all permissions for the particular user. Syntax : user_permission For example:hbase> user_permission hbase> user_permission ‘table1’ |
Monday, October 17, 2016
spark Streaming word count
import org.apache.spark.streaming._
scala> val stc = new StreamingContext(sc, Seconds(5))
stc: org.apache.spark.streaming.StreamingContext = org.apache.spark.streaming.StreamingContext@18f2d752
scala> val lines = stc.socketTextStream("localhost", 9999)
lines: org.apache.spark.streaming.dstream.ReceiverInputDStream[String] = org.apache.spark.streaming.dstream.SocketInputDStream@346995e1
scala> val words = lines.flatMap(_.split(" "))
words: org.apache.spark.streaming.dstream.DStream[String] = org.apache.spark.streaming.dstream.FlatMappedDStream@27058c08
scala> val pairs = words.map(word => (word, 1))
pairs: org.apache.spark.streaming.dstream.DStream[(String, Int)] = org.apache.spark.streaming.dstream.MappedDStream@de5d972
scala> val wordCounts = pairs.reduceByKey(_ + _)
wordCounts: org.apache.spark.streaming.dstream.DStream[(String, Int)] = org.apache.spark.streaming.dstream.ShuffledDStream@409f70ca
scala> wordCounts.print()
scala> wordCounts.foreachRDD { rdd => rdd.foreach(print) }
scala> stc.start()
scala> stc.awaitTermination()
new terminal
[cloudera@quickstart ~]$ nc -lk 9999
hi
hello hello
Tuesday, September 27, 2016
Spark Union,Insection,
Union--Merging two RDDS
scala> val a = sc.parallelize(1 to 5)
scala> val b = sc.parallelize(6 to 10)
scala> val c = a++b
c: org.apache.spark.rdd.RDD[Int] = UnionRDD[2] at $plus$plus at <console>:31
scala> c.collect()
res1: Array[Int] = Array(1, 2, 3, 4, 5, 6, 7, 8, 9, 10)
-------------------
intersection
scala> val a = sc.parallelize(1 to 8)
a: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[3] at parallelize at <console>:27
scala> val b = sc.parallelize(6 to 10)
scala> val c = a++b
c: org.apache.spark.rdd.RDD[Int] = UnionRDD[4] at $plus$plus at <console>:31
scala> c.collect()
res2: Array[Int] = Array(1, 2, 3, 4, 5, 6, 7, 8, 6, 7, 8, 9, 10)
[cloudera@quickstart ~]$ cat abc.txt
Emp1,F,D1
Emp2,M,D2
Emp3,F,D3
Cat
Bat
Mat
[cloudera@quickstart ~]$ cat xyz.txt
Cat
Book
Bat
scala> val a = sc.textFile("/user1/abc.txt")
a: org.apache.spark.rdd.RDD[String] = /user1/abc.txt MapPartitionsRDD[33] at textFile at <console>:27
scala> val b =sc.textFile("/user1/xyz.txt")
b: org.apache.spark.rdd.RDD[String] = /user1/xyz.txt MapPartitionsRDD[35] at textFile at <console>:27
scala> val c = a++b
c: org.apache.spark.rdd.RDD[String] = UnionRDD[36] at $plus$plus at <console>:31
scala> c.collect()
res12: Array[String] = Array(Emp1,F,D1, Emp2,M,D2, Emp3,F,D3, Cat, Bat, Mat, Cat, Book, Bat)
scala> val c = a.intersection(b
| )
c: org.apache.spark.rdd.RDD[String] = MapPartitionsRDD[42] at intersection at <console>:31
scala> c.collect()
res13: Array[String] = Array(Bat, Cat)
----------------------------
Sorting By Key -- It will sort according to key
scala> val a = List((1,"a"),(7,"b"),(5,"F"))
a: List[(Int, String)] = List((1,a), (7,b), (5,F))
scala> val b=sc.parallelize(a);
b: org.apache.spark.rdd.RDD[(Int, String)] = ParallelCollectionRDD[43] at parallelize at <console>:29
scala> val c=b.sortByKey()
c: org.apache.spark.rdd.RDD[(Int, String)] = ShuffledRDD[46] at sortByKey at <console>:31
scala> c.collect()
res14: Array[(Int, String)] = Array((1,a), (5,F), (7,b))
------------
scala> val a = sc.parallelize(2 to 7)
a: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[10] at parallelize at <console>:27
scala> val b = sc.parallelize(5 to 9)
b: org.apache.spark.rdd.RDD[Int] = ParallelCollectionRDD[11] at parallelize at <console>:27
scala> val c = a.intersection(b)
c: org.apache.spark.rdd.RDD[Int] = MapPartitionsRDD[17] at intersection at <console>:31
scala> c.collect()
res6: Array[Int] = Array(6, 7, 5)
--------------------
Sunday, September 4, 2016
Hive index
hive> create table sales(cid int, pid string, amt int)
row format delimited
fields terminated by ',';
OK
Time taken: 11.849 seconds
hive> load data local inpath 'sales' into table sales;
Loading data to table default.sales
Table default.sales stats: [numFiles=1, totalSize=192]
OK
Time taken: 1.142 seconds
hive> select * from sales;
OK
101 p1 1000
102 p1 2000
103 p3 4000
101 p1 1200
101 p3 5000
101 p4 6000
101 p7 9000
102 p2 4000
102 p3 5000
102 p4 6000
103 p1 1000
103 p1 2000
103 p1 4000
102 p2 4000
101 p1 5000
101 p2 3000
Time taken: 0.508 seconds, Fetched: 16 row(s)
hive>
hive> create INDEX cid_index on TABLE
sales(cid)
as 'org.apache.hadoop.hive.ql.index.compact.CompactIndexHandler'
WITH deferred rebuild;
OK
Time taken: 0.351 seconds
hive>
hive> ALTER INDEX cid_index ON sales REBUILD;
hive> show tables;
OK
damp
default__sales_cid_index__
mamp
mytab
ramp
sales
hive> describe default__sales_cid_index__;
OK
cid int
_bucketname string
_offsets array<bigint>
hive> select * from default__sales_cid_index__;
OK
101 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [0,36,48,60,72,168,180]
102 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [12,84,96,108,156]
103 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [24,120,132,144]
[cloudera@quickstart ~]$ cat sales > sales001
hive> load data local inpath 'sales001' into table sales;
Loading data to table default.sales
Table default.sales stats: [numFiles=2, totalSize=384]
OK
Time taken: 0.248 seconds
hive> select * from sales;
OK
101 p1 1000
102 p1 2000
103 p3 4000
101 p1 1200
101 p3 5000
101 p4 6000
101 p7 9000
102 p2 4000
102 p3 5000
102 p4 6000
103 p1 1000
103 p1 2000
103 p1 4000
102 p2 4000
101 p1 5000
101 p2 3000
101 p1 1000
102 p1 2000
103 p3 4000
101 p1 1200
101 p3 5000
101 p4 6000
101 p7 9000
102 p2 4000
102 p3 5000
102 p4 6000
103 p1 1000
103 p1 2000
103 p1 4000
102 p2 4000
101 p1 5000
101 p2 3000
Time taken: 0.073 seconds, Fetched: 32 row(s)
hive>
hive> ALTER INDEX cid_index ON sales REBUILD;
hive> select * from default__sales_cid_index__;
OK
101 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [0,36,48,60,72,168,180]
101 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [0,36,48,60,72,168,180]
102 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [12,84,96,108,156]
102 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [12,84,96,108,156]
103 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [24,120,132,144]
103 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [24,120,132,144]
Time taken: 0.086 seconds, Fetched: 6 row(s)
hive>
[cloudera@quickstart ~]$ cat sales002
101,p1,1000
101,p1,1200
101,p3,5000
101,p4,6000
101,p7,9000
105,p1,9000
105,p4,10000
105,p6,9000
[cloudera@quickstart ~]$
hive> load data local inpath 'sales002' into table sales;
Loading data to table default.sales
Table default.sales stats: [numFiles=3, totalSize=481]
OK
Time taken: 0.233 seconds
hive> select * from sales;
OK
101 p1 1000
102 p1 2000
103 p3 4000
101 p1 1200
101 p3 5000
101 p4 6000
101 p7 9000
102 p2 4000
102 p3 5000
102 p4 6000
103 p1 1000
103 p1 2000
103 p1 4000
102 p2 4000
101 p1 5000
101 p2 3000
101 p1 1000
102 p1 2000
103 p3 4000
101 p1 1200
101 p3 5000
101 p4 6000
101 p7 9000
102 p2 4000
102 p3 5000
102 p4 6000
103 p1 1000
103 p1 2000
103 p1 4000
102 p2 4000
101 p1 5000
101 p2 3000
101 p1 1000
101 p1 1200
101 p3 5000
101 p4 6000
101 p7 9000
105 p1 9000
105 p4 10000
105 p6 9000
Time taken: 0.085 seconds, Fetched: 40 row(s)
hive>
-- in above table , 105 is available only in bucket3(sales002)
hive> select * from default__sales_cid_index__;
OK
101 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [0,36,48,60,72,168,180]
101 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [0,36,48,60,72,168,180]
102 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [12,84,96,108,156]
102 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [12,84,96,108,156]
103 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [24,120,132,144]
103 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [24,120,132,144]
Time taken: 0.074 seconds, Fetched: 6 row(s)
hive>
-- in above output no information about 3rd bucket.
-- bcoz, index is not rebuild.
hive> ALTER INDEX cid_index ON sales REBUILD;
hive> select * from default__sales_cid_index__;
OK
101 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [0,36,48,60,72,168,180]
101 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [0,36,48,60,72,168,180]
101 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales002 [0,12,24,36,48]
102 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [12,84,96,108,156]
102 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [12,84,96,108,156]
103 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales [24,120,132,144]
103 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales001 [24,120,132,144]
105 hdfs://quickstart.cloudera:8020/user/hive/warehouse/sales/sales002 [60,72,85]
Time taken: 0.081 seconds, Fetched: 8 row(s)
hive>
-- after rebuild index, bucket3(sales002) information available.
hive> select * from sales where cid=105;
-- now it reads only bucket3(sales002).
-----------------------------------------------------------------
----------------------------------------------------------------
Hive Bucketing tables and Indexes.
-----------------------------------------------------------------
hive> create table bucks_sales(cid int, pid string,
amt int)
> clustered by (pid)
> into 4 buckets
> row format delimited
> fields terminated by ',';
OK
Time taken: 0.077 seconds
hive>
hive> set hive.enforce.bucketing=true;
hive> insert overwrite table bucks_sales
> select * from sales;
[cloudera@quickstart ~]$ hadoop fs -ls /user/hive/warehouse/bucks_sales
Found 4 items
-rwxrwxrwx 1 cloudera supergroup 73 2016-09-02 11:11 /user/hive/warehouse/bucks_sales/000000_0
-rwxrwxrwx 1 cloudera supergroup 204 2016-09-02 11:11 /user/hive/warehouse/bucks_sales/000001_0
-rwxrwxrwx 1 cloudera supergroup 84 2016-09-02 11:11 /user/hive/warehouse/bucks_sales/000002_0
-rwxrwxrwx 1 cloudera supergroup 120 2016-09-02 11:11 /user/hive/warehouse/bucks_sales/000003_0
[cloudera@quickstart ~]$
-- now data(all rows) divided into 4 buckets.
[cloudera@quickstart ~]$ hadoop fs -cat /user/hive/warehouse/bucks_sales/000000_0
105,p4,10000
101,p4,6000
101,p4,6000
101,p4,6000
102,p4,6000
102,p4,6000
[cloudera@quickstart ~]$ hadoop fs -cat /user/hive/warehouse/bucks_sales/000001_0
101,p1,1000
105,p1,9000
101,p1,1200
101,p1,1000
101,p1,5000
103,p1,4000
103,p1,2000
103,p1,1000
101,p1,1200
102,p1,2000
101,p1,1000
101,p1,5000
103,p1,4000
103,p1,2000
103,p1,1000
101,p1,1200
102,p1,2000
[cloudera@quickstart ~]$ hadoop fs -cat /user/hive/warehouse/bucks_sales/000002_0
102,p2,4000
101,p2,3000
102,p2,4000
105,p6,9000
102,p2,4000
102,p2,4000
101,p2,3000
[cloudera@quickstart ~]$ hadoop fs -cat /user/hive/warehouse/bucks_sales/000003_0
101,p3,5000
102,p3,5000
102,p3,5000
103,p3,4000
101,p7,9000
101,p7,9000
101,p7,9000
101,p3,5000
101,p3,5000
103,p3,4000
[cloudera@quickstart ~]$
-- in above output,
all p4 s available in bucket1 (000000_0)
all p1 s available in bucket2 (000001_0)
all p2 and p6 available in bucket3 (000002_0)
all p3 and p7 available in bucket4 (000003_0)
hive> select * from bucks_sales where pid='p3';
-- to read p3 rows, hive will read all buckets of the table.
-- bcoz, hive does not know in which bucket 'p3' s available.
thats why,
lets create index object on bucks_sales table on column pid.
hive> create index pid_index on table bucks_sales(pid)
as 'org.apache.hadoop.hive.ql.index.compact.CompactIndexHandler'
with deferred rebuild;
hive> show tables;
OK
bucks_sales
damp
default__bucks_sales_pid_index__
default__sales_cid_index__
mamp
mytab
ramp
sales
Time taken: 0.032 seconds, Fetched: 8 row(s)
hive> select * from default__bucks_sales_pid_index__;
OK
Time taken: 0.089 seconds
hive>
-- now index table is empty. bcoz, index is not rebuild(altered).
hive> ALTER INDEX pid_index ON bucks_sales REBUILD;
hive> select * from default__bucks_sales_pid_index__;
OK
p1 hdfs://quickstart.cloudera:8020/user/hive/warehouse/bucks_sales/000001_0 [0,12,24,36,48,60,72,84,96,108,120,132,144,156,168,180,192]
p2 hdfs://quickstart.cloudera:8020/user/hive/warehouse/bucks_sales/000002_0 [0,12,24,48,60,72]
p3 hdfs://quickstart.cloudera:8020/user/hive/warehouse/bucks_sales/000003_0 [0,12,24,36,84,96,108]
p4 hdfs://quickstart.cloudera:8020/user/hive/warehouse/bucks_sales/000000_0 [0,13,25,37,49,61]
p6 hdfs://quickstart.cloudera:8020/user/hive/warehouse/bucks_sales/000002_0 [36]
p7 hdfs://quickstart.cloudera:8020/user/hive/warehouse/bucks_sales/000003_0 [48,60,72]
Time taken: 0.072 seconds, Fetched: 6 row(s)
hive>
hive> select * from bucks_sales where pid='p3';
OK
101 p3 5000
102 p3 5000
102 p3 5000
103 p3 4000
101 p3 5000
101 p3 5000
103 p3 4000
Time taken: 0.078 seconds, Fetched: 7 row(s)
hive>
-- when you ask 'p3' data hive will read only from 4th bucket (000003_0)
Wednesday, August 31, 2016
Mr 3
MR Lab3
grouping By Multiple columns.
ex:
select dno, sex, sum(sal) from emp
group by dno, sex;
DnoSexSalMap.java
--------------------
package mr.analytics;
import java.io.IOException;
import
org.apache.hadoop.io.IntWritable;
import
org.apache.hadoop.io.LongWritable;
import org.apache.hadoop.io.Text;
import
org.apache.hadoop.mapreduce.Mapper;
public class DnoSexSalMap extends
Mapper
<LongWritable,Text,Text,IntWritable>
{
// file : emp
// schema : id,name,sal,sex,dno
// delimiter : "," (comma)
// sample row : 101,amar,20000,m,11
// sex as key, sal as value.
public void map(LongWritable
k,Text v,
Context con)
throws IOException,
InterruptedException
{
String line =
v.toString();
String[] w = line.split(",");
String sex = w[3];
String dno = w[4];
String myKey = dno+"\t"+sex;
int sal =Integer.parseInt(w[2]);
con.write(new Text(myKey),new
IntWritable(sal));
}
}
----------------
Driver8.java
----------------
package mr.analytics;
import
org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.Path;
import
org.apache.hadoop.io.IntWritable;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.mapreduce.Job;
import
org.apache.hadoop.mapreduce.lib.input.F
ileInputFormat;
import
org.apache.hadoop.mapreduce.lib.output.
FileOutputFormat;
public class Driver8
{
public static void main(String
[] args)
throws Exception
{
Configuration c = new
Configuration();
Job j = new Job
(c,"d8");
j.setJarByClass
(Driver8.class);
j.setMapperClass
(DnoSexSalMap.class);
j.setReducerClass
(RedForSum.class);
j.setOutputKeyClass
(Text.class);
j.setOutputValueClass
(IntWritable.class);
Path p1 = new Path
(args[0]); //input
Path p2 = new Path
(args[1]); //output
FileInputFormat.addInputPath(j,p1);
FileOutputFormat.setOutputPath(j, p2);
System.exit(j.waitForCompletion(true) ?
0:1);
}
}
--------------------------
submit:
[training@localhost ~]$ hadoop fs -cat
mrlab/r8/part-r-00000
11 f 25000
11 m 26000
12 f 18000
13 m 19000
______________________________
Spark lab1
spark lab1 : Spark Aggregations : map, flatMap, sc.textFile(), reduceByKey(), groupByKey()
spark Lab1:
___________
[cloudera@quickstart ~]$ cat > comment
i love hadoop
i love spark
i love hadoop and spark
[cloudera@quickstart ~]$ hadoop fs -mkdir spark
[cloudera@quickstart ~]$ hadoop fs -copyFromLocal comment spark
Word Count using spark:
scala> val r1 = sc.textFile("hdfs://quickstart.cloudera/user/cloudera/spark/comment")
scala> r1.collect.foreach(println)
scala> val r2 = r1.map(x => x.split(" "))
scala> val r3 = r2.flatMap(x => x)
instead of writing r2 and r3.
scala> val words = r1.flatMap(x =>
| x.split(" ") )
scala> val wpair = words.map( x =>
| (x,1) )
scala> val wc = wpair.reduceByKey((x,y) => x+y)
scala> wc.collect
scala> val wcres = wc.map( x =>
| x._1+","+x._2 )
scala> wcres.saveAsTextFile("hdfs://quickstart.cloudera/user/cloudera/spark/results2")
[cloudera@quickstart ~]$ cat emp
101,aa,20000,m,11
102,bb,30000,f,12
103,cc,40000,m,11
104,ddd,50000,f,12
105,ee,60000,m,12
106,dd,90000,f,11
[cloudera@quickstart ~]$ hadoop fs -copyFromLocal emp spark
[cloudera@quickstart ~]$
scala> val e1 = sc.textFile("/user/cloudera/spark/emp")
scala> val e2 = e1.map(_.split(","))
scala> val epair = e2.map( x=>
| (x(3), x(2).toInt ) )
scala> val res = epair.reduceByKey(_+_)
res: org.apache.spark.rdd.RDD[(String, Int)] = ShuffledRDD[18] at reduceByKey at <console>:24
scala> res.collect.foreach(println)
(f,170000)
(m,120000)
scala> val resmax = epair.reduceByKey(
| (x,y) => Math.max(x,y))
scala> val resmin = epair.reduceByKey(Math.min(_,_))
scala> resmax.collect.foreach(println)
(f,90000)
(m,60000)
scala> resmin.collect.foreach(println)
(f,30000)
(m,20000)
scala> val grpd = epair.groupByKey()
scala> val resall = grpd.map(x =>
| (x._1, x._2.sum,x._2.size,x._2.max,x._2.min,x._2.sum/x._2.size) )
scala> resall.collect.foreach(println)
Spark lab2
Spark Lab2
scala> val emp = sc.textFile("hdfs://quickstart.cloudera/user/cloudera/spark/emp")
scala> val earray = emp.map(x=> x.split(","))
earray: org.apache.spark.rdd.RDD[Array[String]] = MappedRDD[2] at map at <console>:14
scala> earray.collect
Array[Array[String]] = Array(Array(101, aa, 20000, m, 11), Array(102, bb, 30000, f, 12), Array(103, cc, 40000, m, 11), Array(104, ddd, 50000, f, 12), Array(105, ee, 60000, m, 12), Array(106, dd, 90000, f, 11))
scala> val epair = earray.map( x =>
| (x(4), x(2).toInt))
scala> val ressum = epair.reduceByKey(_+_)
scala> val resmax = epair.reduceByKey(Math.max(_,_))
scala> val resmin = epair.reduceByKey(Math.min(_,_))
scala> ressum.collect.foreach(println)
(12,140000)
(11,150000)
scala> val grpByDno =
epair.groupByKey()
scala> grpByDno.collect
Array[(String, Iterable[Int])] = Array((12,CompactBuffer(30000, 50000, 60000)), (11,CompactBuffer(20000, 40000, 90000)))
scala> val resall = grpByDno.map(x =>
x._1+"\t"+
x._2.sum+"\t"+
x._2.size+"\t"+
x._2.sum/x._2.size+"\t"+
x._2.max+"\t"+
x._2.min )
12 140000 3 46666 60000 30000
11 150000 3 50000 90000 20000
[cloudera@quickstart ~]$ hadoop fs -cat spark/today1/part-00000
12 140000 3 46666 60000 30000
11 150000 3 50000 90000 20000
[cloudera@quickstart ~]$
____________________________________
aggregations by multiple grouping.
ex: equivalant sql/hql query.
select dno, sex , sum(sal) from emp
group by dno, sex;
---
scala> val DnoSexSalPair = earray.map(
| x => ((x(4),x(3)),x(2).toInt) )
scala> DnoSexSalPair.collect.foreach(println)
((11,m),20000)
((12,f),30000)
((11,m),40000)
((12,f),50000)
((12,m),60000)
((11,f),90000)
scala> val rsum = DnoSexSalPair.reduceByKey(_+_)
scala> rsum.collect.foreach(println)
((11,f),90000)
((12,f),80000)
((12,m),60000)
((11,m),60000)
scala> val rs = rsum.map( x =>
x._1._1+"\t"+x._1._2+"\t"+
x._2 )
scala> rs.collect.foreach(println)
11 f 90000
12 f 80000
12 m 60000
11 m 60000
_______________________________________
grouping by multiple columns, and multiple aggregations.
Assignment:
select dno, sex, sum(sal), max(sal) ,
min(sal), count(*), avg(sal)
from emp group by dno, sex;
val grpDnoSex =
DnoSexSalPair.groupByKey();
val r = grpDnoSex.map( x =>
x._1._1+"\t"+
x._1._2+"\t"+
x._2.sum+"\t"+
x._2.max+"\t"+
x._2.min+"\t"+
x._2.size+"\t"+
x._2.sum/x._2.size )
r.collect.foreach(println)
11 f 90000 90000 90000 1 90000
12 f 80000 50000 30000 2 40000
12 m 60000 60000 60000 1 60000
11 m 60000 40000 20000 2 30000
______________________________________
Mr lab1
MR Lab1 : WordCount
[training@localhost ~]$ cat > comment
i love hadoop
i love java
i hate coding
[training@localhost ~]$ hadoop fs -mkdir mrlab
[training@localhost ~]$ hadoop fs -copyFromLocal comment mrlab
[training@localhost ~]$ hadoop fs -cat mrlab/comment
i love hadoop
i love java
i hate coding
[training@localhost ~]$
___________________________
Eclipse steps :
create java project:
file--> new --> java project
ex: MyMr
configure jar files.
MyMr--> src --> Build Path --> Configure build Path
---> Libraries ---> add external jar files.
select folloing jars.
/usr/lib/hadoop-*.*/hadoop-core.jar
/usr/lib/hadoop-*.*/lib/commons-cli-*.*.jar
create package :
MyMr --> src --> new --> package
ex: mr.analytics
create java class :
MyMr --> src --> mr.analytics --> new --> class
ex: WordMap
__________________________________
WordMap.java:
is a mapper program, which writes word as key and 1 as value.
----------------------
package mr.analytics;
import java.io.IOException;
import org.apache.hadoop.io.IntWritable;
import org.apache.hadoop.io.LongWritable;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.mapreduce.Mapper;
import java.util.StringTokenizer;
public class WordMap extends Mapper
<LongWritable,Text,Text,IntWritable>
{
// i love hadoop
public void map(LongWritable k,Text v,
Context con)
throws IOException, InterruptedException
{
String line = v.toString();
StringTokenizer t = new StringTokenizer(line);
while(t.hasMoreTokens())
{
String word = t.nextToken();
con.write(new Text(word),new IntWritable(1));
}
}
}
---------------------
RedForSum.java
is Reducer program, which will give sum aggregation.
----------------------------
package mr.analytics;
import java.io.IOException;
import org.apache.hadoop.io.IntWritable;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.mapreduce.Reducer;
public class RedForSum extends Reducer
<Text,IntWritable,Text,IntWritable>
{
// i <1,1,1>
public void reduce(Text k,Iterable<IntWritable> vlist,
Context con)
throws IOException, InterruptedException
{
int tot=0;
for(IntWritable v: vlist)
tot+=v.get();
con.write(k, new IntWritable(tot));
}
}
---------------------------
Driver1.java
is Driver program, to call mapper and reducer
-----------------
package mr.analytics;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.IntWritable;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.mapreduce.Job;
import org.apache.hadoop.mapreduce.lib.input.FileInputFormat;
import org.apache.hadoop.mapreduce.lib.output.FileOutputFormat;
public class Driver1
{
public static void main(String[] args)
throws Exception
{
Configuration c = new Configuration();
Job j = new Job(c,"MyFirst");
j.setJarByClass(Driver1.class);
j.setMapperClass(WordMap.class);
j.setReducerClass(RedForSum.class);
j.setOutputKeyClass(Text.class);
j.setOutputValueClass(IntWritable.class);
Path p1 = new Path(args[0]); //input
Path p2 = new Path(args[1]); //output
FileInputFormat.addInputPath(j,p1);
FileOutputFormat.setOutputPath(j, p2);
System.exit(j.waitForCompletion(true) ? 0:1);
}
}
---------------------
Now export all these 3 classes into jar file.
MyMr --> export --> jar -->java jar
ex: /home/training/Desktop/myapp.jar
-------------------------------
submitting mr job:
[training@localhost ~]$ hadoop jar Desktop/myapp.jar \
> mr.analytics.Driver1 \
> mrlab/comment \
> mrlab/res1
[training@localhost ~]$ hadoop fs -ls mrlab/res1
Found 3 items
-rw-r--r-- 1 training supergroup 0 2016-08-30 05:27
/user/training/mrlab/res1/_SUCCESS
drwxr-xr-x - training supergroup 0 2016-08-30 05:26
/user/training/mrlab/res1/_logs
-rw-r--r-- 1 training supergroup 58 2016-08-30 05:27
/user/training/mrlab/res1/part-r-00000
[training@localhost ~]$ hadoop fs -cat mrlab/res1/part-r-00000
coding 1
hadoop 1
hate 1
i 3
java 1
love 2
Spark lab1
spark lab1 : Spark Aggregations : map, flatMap, sc.textFile(), reduceByKey(), groupByKey()
spark Lab1:
___________
[cloudera@quickstart ~]$ cat > comment
i love hadoop
i love spark
i love hadoop and spark
[cloudera@quickstart ~]$ hadoop fs -mkdir spark
[cloudera@quickstart ~]$ hadoop fs -copyFromLocal comment spark
Word Count using spark:
scala> val r1 = sc.textFile("hdfs://quickstart.cloudera/user/cloudera/spark/comment")
scala> r1.collect.foreach(println)
scala> val r2 = r1.map(x => x.split(" "))
scala> val r3 = r2.flatMap(x => x)
instead of writing r2 and r3.
scala> val words = r1.flatMap(x =>
| x.split(" ") )
scala> val wpair = words.map( x =>
| (x,1) )
scala> val wc = wpair.reduceByKey((x,y) => x+y)
scala> wc.collect
scala> val wcres = wc.map( x =>
| x._1+","+x._2 )
scala> wcres.saveAsTextFile("hdfs://quickstart.cloudera/user/cloudera/spark/results2")
[cloudera@quickstart ~]$ cat emp
101,aa,20000,m,11
102,bb,30000,f,12
103,cc,40000,m,11
104,ddd,50000,f,12
105,ee,60000,m,12
106,dd,90000,f,11
[cloudera@quickstart ~]$ hadoop fs -copyFromLocal emp spark
[cloudera@quickstart ~]$
scala> val e1 = sc.textFile("/user/cloudera/spark/emp")
scala> val e2 = e1.map(_.split(","))
scala> val epair = e2.map( x=>
| (x(3), x(2).toInt ) )
scala> val res = epair.reduceByKey(_+_)
res: org.apache.spark.rdd.RDD[(String, Int)] = ShuffledRDD[18] at reduceByKey at <console>:24
scala> res.collect.foreach(println)
(f,170000)
(m,120000)
scala> val resmax = epair.reduceByKey(
| (x,y) => Math.max(x,y))
scala> val resmin = epair.reduceByKey(Math.min(_,_))
scala> resmax.collect.foreach(println)
(f,90000)
(m,60000)
scala> resmin.collect.foreach(println)
(f,30000)
(m,20000)
scala> val grpd = epair.groupByKey()
scala> val resall = grpd.map(x =>
| (x._1, x._2.sum,x._2.size,x._2.max,x._2.min,x._2.sum/x._2.size) )
scala> resall.collect.foreach(println)
Monday, August 29, 2016
Spark Word count
[cloudera@quickstart ~]$ cat as.txt
i love hadoop
i love spark
o love hadoop and spark
scala> val rdd = sc.textFile("/user1/as.txt")
rdd: org.apache.spark.rdd.RDD[String] = /user1/as.txt MapPartitionsRDD[42] at textFile at <console>:31
scala> val rdd1 =rdd.flatMap(x=>x.split(" "))
rdd1: org.apache.spark.rdd.RDD[String] = MapPartitionsRDD[43] at flatMap at <console>:33
scala> val rdd2 =rdd1.map(x=>(x,1))
rdd2: org.apache.spark.rdd.RDD[(String, Int)] = MapPartitionsRDD[44] at map at <console>:35
scala> val rdd3 = rdd2.reduceByKey(_+_)
rdd3: org.apache.spark.rdd.RDD[(String, Int)] = ShuffledRDD[9] at reduceByKey at <console>:33
scala> val rdd4 = rdd3.map(x => x._1 + " " +x._2).collect()
rdd4: Array[String] = Array(love 3, spark 2, hadoop 2, i 2, o 1, and 1)
Tuesday, August 23, 2016
spark json
[cloudera@quickstart ~]$ cat a.json
{"name":"shr","age":23,"loc":"hyd"}
{"name":"kumar","age" :24,"loc":"del"}
scala> import org.apache.spark.sql.SQLContext
import org.apache.spark.sql.SQLContext
scala> val sql= new SQLContext(sc)
sql: org.apache.spark.sql.SQLContext = org.apache.spark.sql.SQLContext@410438f6
scala> val df =sql.read.json("/user1/a.json")
df: org.apache.spark.sql.DataFrame = [age: bigint, loc: string, name: string]
scala> df.show()
+---+---+-----+
|age|loc| name|
+---+---+-----+
| 23|hyd| shr|
| 24|del|kumar|
+---+---+-----+
scala> df.select("name").show()
+-----+
| name|
+-----+
| shr|
|kumar|
+-----+
scala> df.select(df("age")+1).show()
+---------+
|(age + 1)|
+---------+
| 24|
| 25|
+---------+
scala> df.first()
res10: org.apache.spark.sql.Row = [23,hyd,shr]
scala> df.take(1)
res12: Array[org.apache.spark.sql.Row] = Array([23,hyd,shr])
scala> df.take(2)
res13: Array[org.apache.spark.sql.Row] = Array([23,hyd,shr], [24,del,kumar])
scala> df.sort().show()
+---+---+-----+
|age|loc| name|
+---+---+-----+
| 23|hyd| shr|
| 24|del|kumar|
+---+---+-----+
scala> df.filter(df("age")>23).show()
+---+---+-----+
|age|loc| name|
+---+---+-----+
| 24|del|kumar|
+---+---+-----+
scala> df.describe().show()
+-------+------------------+
|summary| age|
+-------+------------------+
| count| 2|
| mean| 23.5|
| stddev|0.7071067811865476|
| min| 23|
| max| 24|
+-------+------------------+
scala> df.agg(avg("age")).show()
+--------+
|avg(age)|
+--------+
| 23.5|
+--------+
{"name":"kumar","age" :24,"loc":"del","sal":4000,"sex":"f"}
{"name":"aaaa","age" :24,"loc":"wgl","sal":5000,"sex":"m"}
{"name":"bbbb","age" :24,"loc":"del","sal":1000,"sex":"m"}
{"name":"ccccc","age" :24,"loc":"pune","sal":5000,"sex":"f"}
{"name":"ddddddd","age" :24,"loc":"del","sal":3000,"sex":"m"}
{"name":"eeeee","age" :24,"loc":"hyd","sal":2000,"sex":"f"}
[cloudera@quickstart ~]$ vi a.json
[cloudera@quickstart ~]$ cat a.json
{"name":"kumar","age" :24,"loc":"del","sal":4000,"sex":"f"}
{"name":"aaaa","age" :34,"loc":"wgl","sal":5000,"sex":"m"}
{"name":"bbbb","age" :25,"loc":"del","sal":1000,"sex":"m"}
{"name":"ccccc","age" :21,"loc":"pune","sal":5000,"sex":"f"}
{"name":"ddddddd","age" :44,"loc":"del","sal":3000,"sex":"m"}
{"name":"eeeee","age" :30,"loc":"hyd","sal":2000,"sex":"f"}
[cloudera@quickstart ~]$ hadoop fs -put a.json /user1/
scala> import org.apache.spark.sql.SQLContext
import org.apache.spark.sql.SQLContext
scala> val sql= new SQLContext(sc)
sql: org.apache.spark.sql.SQLContext = org.apache.spark.sql.SQLContext@60ae0f04
scala> val df =sql.read.json("/user1/a.json")
df: org.apache.spark.sql.DataFrame = [age: bigint, loc: string, name: string, sal: bigint, sex: string]
scala> df.show()
+---+----+-------+----+---+
|age| loc| name| sal|sex|
+---+----+-------+----+---+
| 24| del| kumar|4000| f|
| 34| wgl| aaaa|5000| m|
| 25| del| bbbb|1000| m|
| 21|pune| ccccc|5000| f|
| 44| del|ddddddd|3000| m|
| 30| hyd| eeeee|2000| f|
+---+----+-------+----+---+
scala> df.select("age").show()
+---+
|age|
+---+
| 24|
| 34|
| 25|
| 21|
| 44|
| 30|
+---+
scala> df.filter(df("age")<30).show()
+---+----+-----+----+---+
|age| loc| name| sal|sex|
+---+----+-----+----+---+
| 24| del|kumar|4000| f|
| 25| del| bbbb|1000| m|
| 21|pune|ccccc|5000| f|
+---+----+-----+----+---+
scala> df.groupBy("sex").count().show()
+---+-----+
|sex|count|
+---+-----+
| f| 3|
| m| 3|
+---+-----+
scala> df.where(df("age")<25).show()
+---+----+-----+----+---+
|age| loc| name| sal|sex|
+---+----+-----+----+---+
| 24| del|kumar|4000| f|
| 21|pune|ccccc|5000| f|
+---+----+-----+----+---+
scala> val e= df.toJSON
e: org.apache.spark.rdd.RDD[String] = MapPartitionsRDD[269] at toJSON at <console>:38
scala> e.collect()
res73: Array[String] = Array({"age":24,"loc":"del","name":"kumar","sal":4000,"sex":"f"}, {"age":34,"loc":"wgl","name":"aaaa","sal":5000,"sex":"m"}, {"age":25,"loc":"del","name":"bbbb","sal":1000,"sex":"m"}, {"age":21,"loc":"pune","name":"ccccc","sal":5000,"sex":"f"}, {"age":44,"loc":"del","name":"ddddddd","sal":3000,"sex":"m"}, {"age":30,"loc":"hyd","name":"eeeee","sal":2000,"sex":"f"})
scala> df.describe().show()
+-------+------------------+------------------+
|summary| age| sal|
+-------+------------------+------------------+
| count| 6| 6|
| mean|29.666666666666668|3333.3333333333335|
| stddev| 8.406346808612327|1632.9931618554522|
| min| 21| 1000|
| max| 44| 5000|
+-------+------------------+------------------+
scala> df.agg(max("sal")).show()
+--------+
|max(sal)|
+--------+
| 5000|
+--------+
scala> df.sort().show()
+---+----+-------+----+---+
|age| loc| name| sal|sex|
+---+----+-------+----+---+
| 44| del|ddddddd|3000| m|
| 30| hyd| eeeee|2000| f|
| 24| del| kumar|4000| f|
| 34| wgl| aaaa|5000| m|
| 25| del| bbbb|1000| m|
| 21|pune| ccccc|5000| f|
+---+----+-------+----+---+
scala> df.sort("sal").show()
+---+----+-------+----+---+
|age| loc| name| sal|sex|
+---+----+-------+----+---+
| 25| del| bbbb|1000| m|
| 30| hyd| eeeee|2000| f|
| 44| del|ddddddd|3000| m|
| 24| del| kumar|4000| f|
| 34| wgl| aaaa|5000| m|
| 21|pune| ccccc|5000| f|
+---+----+-------+----+---+
scala> df.take(2)
res62: Array[org.apache.spark.sql.Row] = Array([24,del,kumar,4000,f], [34,wgl,aaaa,5000,m])
scala> df.first()
res63: org.apache.spark.sql.Row = [24,del,kumar,4000,f]
scala> df.head(3)
res69: Array[org.apache.spark.sql.Row] = Array([24,del,kumar,4000,f], [34,wgl,aaaa,5000,m], [25,del,bbbb,1000,m])
scala> df.orderBy("age").show()
+---+----+-------+----+---+
|age| loc| name| sal|sex|
+---+----+-------+----+---+
| 21|pune| ccccc|5000| f|
| 24| del| kumar|4000| f|
| 25| del| bbbb|1000| m|
| 30| hyd| eeeee|2000| f|
| 34| wgl| aaaa|5000| m|
| 44| del|ddddddd|3000| m|
+---+----+-------+----+---+
scala> df.orderBy("sal").show()
+---+----+-------+----+---+
|age| loc| name| sal|sex|
+---+----+-------+----+---+
| 25| del| bbbb|1000| m|
| 30| hyd| eeeee|2000| f|
| 44| del|ddddddd|3000| m|
| 24| del| kumar|4000| f|
| 34| wgl| aaaa|5000| m|
| 21|pune| ccccc|5000| f|
+---+----+-------+----+---+